Revolutionizing Biology: The Impact of AlphaFold on Protein Research
Written on
Chapter 1: The AlphaFold Revolution
AlphaFold from DeepMind has created a seismic shift in the field of biology. This groundbreaking AI tool is reshaping how researchers approach protein structure prediction.
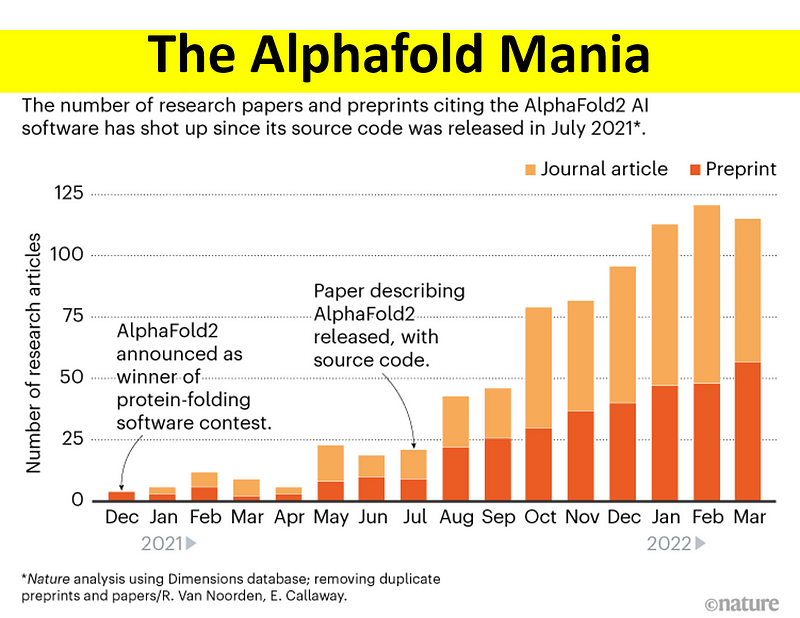
The publication of the AlphaFold2 paper, along with its source code, in July 2021 marked a pivotal moment. The upward trend in related articles since then underscores its significance. For those unfamiliar, AlphaFold2 (AF2) is an AI framework that predicts the three-dimensional configuration of proteins based solely on their amino acid sequences.
Previously, this challenge was met with various heuristic methods and first-principles programming until DeepMind demonstrated a more effective approach. Instead of manually coding the specific roles of each amino acid, they trained the AI to learn the underlying principles by analyzing vast datasets of protein structures. The result was a system that outperformed traditional methods.
Today, AlphaFold has become foundational in proteomics and drug discovery. Every drug interacts with specific proteins, and understanding these structures is crucial for discerning how drugs, genes, or even cancer cells engage with them. AlphaFold not only accelerates research but also opens avenues previously deemed impossible, enhancing the vibrancy of scientific inquiry.
Despite its transformative potential, AlphaFold is not without limitations; some of its predictions may lack reliability. Nevertheless, we now operate in a distinct era characterized by the profound influence of AlphaFold.
Looking ahead, DeepMind plans to unveil over 100 million structural predictions this year. This ambitious endeavor represents nearly half of all known proteins and vastly exceeds the number of experimentally determined structures in the Protein Data Bank (PDB).
The future holds the promise of discovering new proteins with unprecedented and beneficial properties.